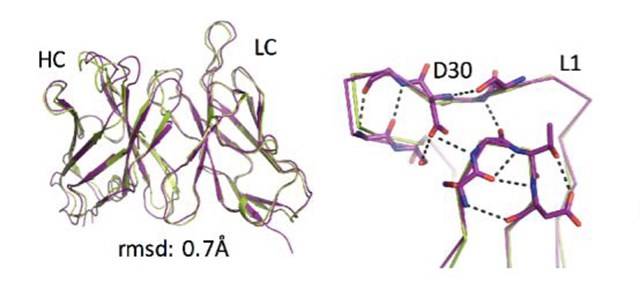
Computer designs (lime green) are compared with experimental structures (purple) at the atomic level, revealing atomic accuracy in overall structure (left) and in loop regions (right)
Including “non-ideal” components in the design may be key to functional artificial proteins
The idea of proteins that can be designed on computers for specific functions has been a cutting-edge concept that has stubbornly remained “in the future.” New research at the Weizmann Institute of Science may bring that future a bit closer. By going back to nature’s drawing board – evolution – the scientists have created new proteins, based on “existing natural parts,” that carry out their intended function with flying colors. These findings were reported in the Proceedings of the National Academy of Sciences (PNAS).
The research was conducted by Dr. Sarel Fleishman and members of his group in the Department of Biomolecular Sciences, including research students Dror Baran, Maria Gabriele Pszolla, and Gideon Lapidoth. In Dr. Fleishman’s lab, proteins are designed with computer-based programs that enable them to generate new structures – for example, antibodies or enzymes – that do not exist in nature. If the team wants a protein that will perform a specific action – say, bind to another protein or carry out a chemical reaction – they can compute, from beginning to end, the genetic sequence that will line up amino acids in the proper order and cause the protein to fold into the correct three-dimensional shape. Such proteins could, in theory, usher in a new age of custom-designed drugs and catalysts, but the challenges of planning of large biological molecules are immense.

The team asked a few simple questions: “What does a natural enzyme or antibody have that the artificial proteins don’t? Why are two structures with similar makeup so different when it comes to the way they perform inside a biological system?”
The group focused their attention on some parts of natural antibodies or enzymes that start from scratch and don’t make it into the computer designs; in particular, structures called “loops,” which are inherently unstable and “non-ideal” and, therefore, challenging when it comes to computational prediction. These “non-ideal” loops can often be found at the very center of the active regions – those that recognize a target or bind to or cleave another molecule.
To incorporate these elements, the researchers decided to design a functioning antibody from existing parts, rather than building one from scratch. They broke the structures found in natural antibodies down into segments, including the loops and other supporting features. In effect, the researchers tinkered with ready-made parts, similar to the way evolution works.

Learning the rules of antibody design: Flaws in designed insulin-targeting antibodies (insets) are addressed through five successive design-build-test rounds of algorithm “evolution,” raising the fraction of designed antibodies stably expressed by cells from near 0% to 100%
Natural evolution is, of course, a very slow process: A new family of antibodies can be tens of millions of years in the making. So the researchers went back to the computerized planning process, this time armed with their new insight. The new designs were tested experimentally in the lab, a few dozen new antibodies at a time. Initially, the designs performed poorly, but through five design-build-test cycles, the team uncovered some general rules for designing antibodies. In essence, they created a sort of symbiotic evolution – the design programs were evolving along with the experimental tests, each pushing the other forward. To demonstrate the feasibility of this concept, the team created artificial antibodies that targeted insulin, characterizing these molecules down to the resolution of single atoms.
In future experiments, the scientists plan to design artificial antibodies modeled on those of camels and llamas. Why these particular animals? If a human antibody, or one from any number of common animals, has 200 amino acids, antibodies in camels and llamas are made of just 100 – and yet are stable and effective. This could make the design and production of artificial antibodies for human conditions more efficient, and might have relevance for designing new diagnostics and therapeutics.
Dr. Sarel-Jacob Fleishman’s research is supported by the Henri Gutwirth Fund for Research; the Rothschild Caesarea Foundation; Sam Switzer, Canada; Anne Christopoulos, Canada; and Carolyn Hewitt, Canada. Dr. Fleishman is the incumbent of the Martha S. Sagon Career Development Chair.